Achievements
The isoleucine residue at position 35 of ALS mutant SOD1 plays a key role in intracellular aggregation
A paper submitted as the first author by Dr. Araki Asai, who graduated from our School of Medicine in March this year and received the President's Award for the Research Physician Course, was published in the international journal "Molecular Neurobiology" on July 26, 2024. Dr. Asai has been conducting research with Professor Noriko Fujiwara in the Department of Biochemistry Department for four years as a research student. In this study, it was revealed that isoleucine (I35), the 35th amino acid residue of ALS mutant SOD1, plays an important role in the formation of SOD1 aggregates.
Research Background
In neurodegenerative diseases including amyotrophic lateral sclerosis (ALS), aggregation and misfolding of the causative protein are thought to be the cause of the pathology. Cu/Zn-superoxide dismutase (SOD1) is an antioxidant enzyme that scavenges superoxide, and it has been revealed that mutations in the SOD1 gene cause familial amyotrophic lateral sclerosis (ALS). SOD1 functions as a dimer with two subunits consisting of 153 amino acids. More than 200 point mutations and ALS mutations with C-terminal deletions have been found so far. It has been reported that such ALS mutant SOD1 has a more unstable structure and is more prone to aggregation than wild-type SOD1, suggesting a relationship with the onset of ALS. However, the mechanism of ALS onset has not yet been elucidated, and no effective treatment has yet been found.
We therefore hypothesized that elucidating the mechanism of SOD1 aggregate formation in cells would lead to elucidation of the mechanism of ALS onset, and fused enhanced green fluorescent protein (EGFP) to the C-terminus of SOD1 and expressed it in HEK293 cells. However, ALS mutant SOD1 did not form many aggregates, and we were unable to find any difference from wild-type SOD1.
Research Results
In this study, we found that when an expression system in which BioID2 was fused to the N-terminus of SOD1-EGFP was used, intracellular aggregate formation was significantly increased with ALS mutant SOD1 compared to wild-type SOD1. Using this expression system, we sequentially deleted the C-terminus of SOD1 and found that aggregates were not formed when the amino acid sequence was shorter than the N-terminus partial sequence of SOD1, "1-34," but was formed when the amino acid sequence was longer than "1-35" (deletion mutations such as 1-120 and 1-140). Furthermore, when the 35th amino acid residue of ALS mutant SOD1, isoleucine (I35), was replaced with serine (S), a hydrophilic amino acid residue, aggregate formation was significantly suppressed.
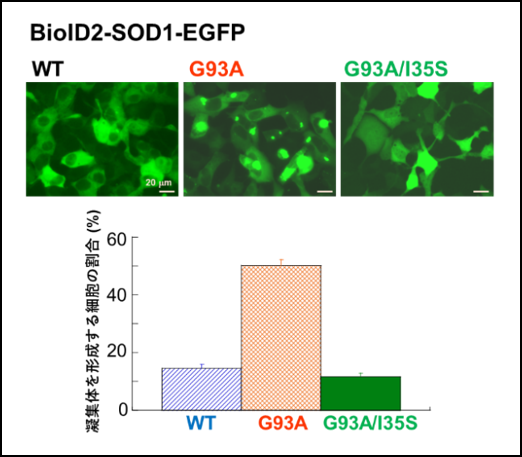
Figure 1 shows that aggregate formation is increased in the ALS mutant SOD1 G93A (the 93rd amino acid is mutated from glycine to alanine) compared to wild-type (WT) SOD1, and that aggregate formation is suppressed in G93A/I35S, in which I35 is mutated to serine.
These results demonstrated that the 35th amino acid residue, isoleucine (I35), of SOD1 plays an important role in aggregate formation.
Future challenges and prospects
So far, no familial ALS patients with I35 mutations have been reported. In other words, all ALS patients, including those with sporadic ALS, have SOD1 with I35, which is important for aggregate formation. In the future, we will need to confirm in ALS model animals whether I35-mediated SOD1 aggregate formation is involved in ALS onset. If it becomes clear that aggregate formation is directly involved in ALS onset, we hope that this may pave the way for drug development that targets I35 as a new target.
Publication information
Journal
Molecular Neurobiology(IMPACT FACTOR : 4.6)
Title of the paper
The Ile35 Residue of the ALS-Associated Mutant SOD1 Plays a Crucial Role in the Intracellular Aggregation of the Molecule
The Ile35 Residue of the ALS-Associated Mutant SOD1 Plays a Crucial Role in the Intracellular Aggregation of the Molecule
Author(s) of the paper
Shinki Asai*1, Anzu Yano*1, Tomoyuki Higashino*1, Daisaku Yoshihara*1, Haruhiko Sakiyama*1, Hironobu Eguchi*1
Kazuaki Fukushima*2, Keiichiro Suzuki*1, Noriko Fujiwara*1
*1 Hyogo Medical University ・ School of Medicine ・ Department of Biochemistry Lecture
*2 Hyogo Medical University ・ School of Medicine ・ Chair of Chemistry
Source of research funds
Research Support Grant for Research Doctor Course (Asai Araki)
Grant-in-Aid for Scientific Research C (22K11870 Representative: Noriko Fujiwara)
Japan Association for Applied Enzyme Research Grant (Noriko Fujiwara)
"Initiative to Realize a Diversity Research Environment (Characteristics-Based)" by the Ministry of Education, Culture, Sports, Science and Technology's Science and Technology Human Resource Development Subsidy Program (Hyogo Medical University, 2023 English Paper Submission Support) (Noriko Fujiwara)