About the Corporation
Dysfunction of cytotoxic T cells may be involved in the worsening of COVID-19 symptoms - Towards the creation of a "universal vaccine" that is effective against new mutant viruses -
A research group led by Professor Satoshi Hyogo Medical University, Chair of School of Medicine Department of Microbiology, and Lecturer Hideki Ogura, Professor Junichi Hirata, Chair of the Department of Department of Emergency Disaster and Critical Care Medicine, and Lecturer Kunihiro Shirai, and Visiting Professor Yoshio Takesue, Department of Infection Prevention and Control at Hyogo College of Medicine, in collaboration with Kyowakai Medical Corporation (Kawanishi City, Hyogo Prefecture; Chair of the Board of Trustees: Toru Kitagawa), the University of Tokyo (Bunkyo Ward, Tokyo; President: Teruo Fujii), Osaka University (Suita City, Osaka Prefecture; President: Shojiro Nishio), and Kumamoto University (Kumamoto City, Kumamoto Prefecture; President: Hisao Ogawa), conducted surveys and analyses of patients who had recovered from COVID-19 and identified antigenic peptides recognized by T cells, which play an important role in eliminating the virus.
A paper on the results of this research was published in the academic journal Nature Communications at 7:00 p.m. on December 16, 2022 (Japan time).
[Key points of this research]
While research into the novel coronavirus has primarily focused on antibodies (humoral immunity), another important mechanism for eliminating the virus is cellular immunity. Regarding the characteristics of virus-specific cytotoxic T lymphocytes (CTLs) related to the severity of COVID-19, which has yet to be fully investigated, we explored the antigenic sites recognized by CTLs derived from Japanese convalescent COVID-19 patients.
●As a result, in patients who recovered from severe COVID-19 and had the human leukocyte antigen (HLA-A*24:02), which is predominant in Japanese people, CTLs that recognize "M198-206," which is common to different novel coronavirus variants, were undifferentiated and exhausted. This revealed the possibility that dysfunction of CTLs that recognize "M198-206" may lead to the worsening of COVID-19 disease.
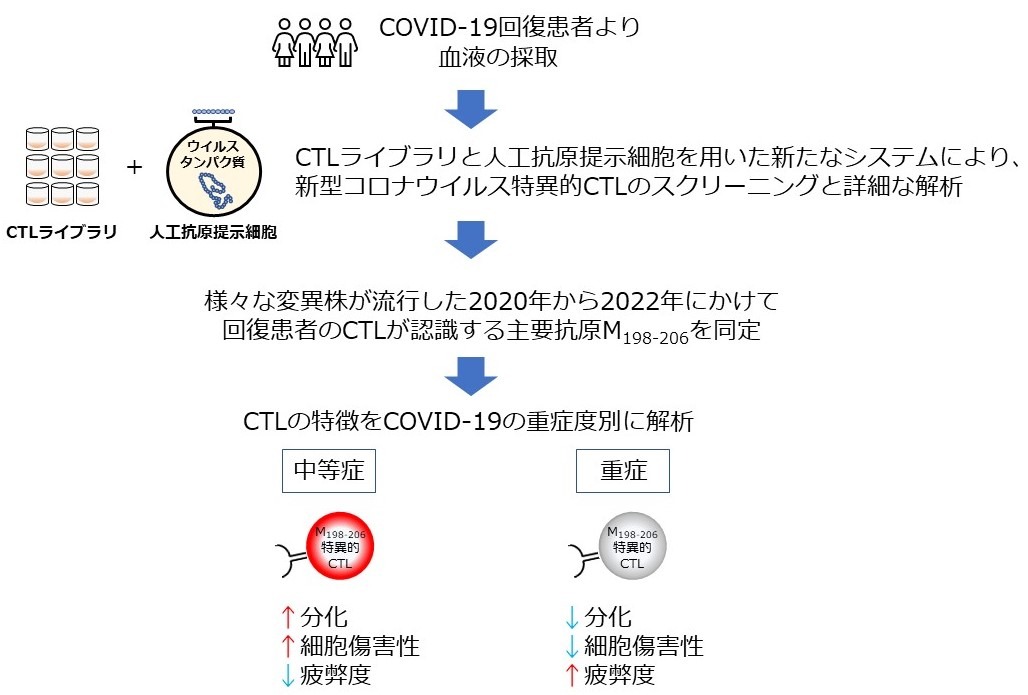
Research background and aims
The relationship between abnormalities in CTLs that recognize the novel coronavirus and the severity of COVID-19 remains unclear
The novel coronavirus continues to mutate, and COVID-19 shows no signs of ending. The mortality rate of COVID-19 is high among the elderly, so continued efforts are needed. Abnormalities in innate immunity, such as type I IFN, have been identified as a cause of severe COVID-19. Abnormalities in adaptive immunity, such as a decrease in the overall number of T cells, have also been identified. However, among CTLs that work on the front lines of viral elimination, the relationship between the severity of the disease and specific CTLs targeting the novel coronavirus has been insufficiently analyzed, as the primary target site (epitope) within the novel coronavirus is not well understood.
Detailed investigation of the characteristics of virus-specific CTLs that lead to aggravation is extremely important in obtaining clues for treatment strategies. For example, if abnormalities in the expression of a certain gene in the CTLs of patients with severe disease are identified, drug discovery strategies targeting that gene will emerge. Furthermore, by examining the relationship between CTL characteristics and aggravation, it will be possible to identify CTLs that contributed to recovery, and their epitopes will provide clues for the creation of safer vaccines.
Therefore, Hyogo Medical University, Kyowakai, the University of Tokyo, Osaka University, and Kumamoto University collaborated on the following activities with the aim of "clarifying the importance of CTLs in recovery from COVID-19 and obtaining hints for new treatment strategies."
① By constructing a new screening system for specific CTLs that recognize the new coronavirus, we will identify specific CTLs and their epitopes that are abundant in recovered patients.
② Using this epitope, we investigated the characteristics of CTLs that recognize the new coronavirus in recovered COVID-19 patients with different clinical courses.
Implementation methods and results
The researchers found that "COVID-19-specific CTLs in patients recovering from severe COVID-19 show signs of dysfunction and exhaustion," and discovered that abnormalities in CTLs that recognize "M198-206," a common amino acid sequence among different COVID-19 strains, may lead to the worsening of COVID-19 severity.
We analyzed CTLs that recognize the novel coronavirus using a new screening system that uses a CTL library and artificial antigen-presenting cells in patients who received treatment for COVID-19 and recovered from the disease at Hyogo Medical University of Medicine and Kyowakai Medical Corporation between 2020 and 2022.
<Implementation method>
A CTL library was created from the blood of patients who had recovered from COVID-19, and novel coronavirus-specific CTLs that reacted with artificial antigen-presenting cells expressing novel coronavirus proteins were searched for, and analyses were performed, including the discovery of epitopes recognized by these CTLs, cytokine profiling, cytotoxicity assays, tetramer staining, and single-cell RNA sequencing.
As a result, the following became clear:
① In recovered Japanese patients with HLA-A*24:02, which is the most common type in Japanese people, the M protein of the new coronavirus(※)CTLs that recognize the virus will be detected in large numbers from the end of 2020 to the beginning of 2022.
*This protein is the most abundant in virus particles and is essential for virus formation, functioning as a scaffold for virus particle formation. It is highly conserved within the β-CoV genus.
② The major epitope in the M protein is M198-206, and this region is conserved in all virus strains that have circulated to date.
3) M198-206-specific CTLs actually suppress the proliferation of the novel coronavirus in alveolar epithelial cells in vitro.
④ M198-206-specific CTLs from recovered patients with severe COVID-19 are less differentiated and more exhausted than CTLs from recovered patients with moderate COVID-19.
⑤ Multiple common T cell receptors were found in M198-206-specific CTLs from recovered patients.
From the above findings (1) to (5), it became clear that "in Japanese people, who have a high frequency of HLA-A*24:02, the function of CTLs that recognize the novel coronavirus M protein may be important in preventing the worsening of COVID-19." The CTL epitope, M198-206, is completely conserved among virus strains, and therefore is thought to be the basis for the development of a CTL-inducing vaccine that is effective against virus strains that are expected to emerge in the future.
Future use and development
Creating a "universal vaccine" that is effective against viruses yet to be discovered
The importance of CTLs in eliminating the virus is clear in COVID-19, but the characteristics of CTLs related to the severity of the disease have not been fully analyzed. To investigate these characteristics, we explored the antigenic sites recognized by CTLs from Japanese patients with COVID-19 in the convalescent stage and succeeded in identifying the sites that many people actually react to with HLA types common in Japanese people.
Furthermore, it was found that some T cell receptors of CTLs against the novel coronavirus are shared between patients. However, further analysis is required to explore the causes of CTL dysfunction in severely ill patients (and to apply this to patients with other HLA types). Furthermore, the T cell epitopes identified in this study could potentially be used as vaccines to induce CTLs, which could contribute to the development of treatments for vaccine-resistant individuals who are less likely to induce antibody production or who have a weak CTL response. It is also expected that T cell epitopes could be used to detect CTLs and monitor cellular immunity.
[Paper title]
Dysfunctional Sars-CoV-2 M protein-specific cytotoxic T lymphocytes in patients recovering from severe COVID-19
[Paper author name]
Hideki Ogura(1*), Jin Gohda(2), Xiuyuan Lu(3), Mizuki Yamamoto(2), Yoshio Takesue(4,5), Aoi Son(1), Sadayuki Doi(6), Kazuyuki Matsushita(7), Fumitaka Isobe(8), Yoshihiro Fukuda(9), Tai-Ping Huang(10), Takamasa Ueno(11), Naomi Mambo(12), Hiromoto Murakami(12), Yasushi Kawaguchi(2,13), Jun-ichiro Inoue(14), Kunihiro Shirai(12), Sho Yamasaki(3,15,16,17), Jun-Ichi Hirata(12) and Satoshi Ishido(1*)
(1)Department of Microbiology, Hyogo Medical University, Hyogo, Japan.
(2)Research Center for Asian Infectious Disease, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan.
(3)Laboratory of Molecular Immunology, Immunology Frontier Research Center, Osaka University, Suita, Japan.
(4)Department of Infection Control and Prevention, Hyogo Medical University, Hyogo, Japan.
(5)Tokoname City Hospital, Aichi, Japan.
(6) Kawanishi City Hospital, Hyogo, Japan.
(7)Kyoritsu Hospital, Hyogo, Japan.
(8) Kyowa Marina Hospital/Wellhouse Nishinomiya, Hyogo, Japan.
(9)Dainikyoritsu Hospital, Hyogo, Japan.
(10)Kyoritsu Onsen Hospital, Hyogo, Japan.
(11)Joint Research Center for Human Retrovirus Infection, Kumamoto University, Kumamoto, Japan.
(12)Department of Emergency and Critical Care Medicine, Hyogo Medical University, Hyogo, Japan.
(13)Division of Molecular Virology, Department of Microbiology and Immunology, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan.
(14)Research Platform Office, The Institute of Medical Science, The University of Tokyo, Tokyo, Japan.
(15)Department of Molecular Immunology, Research Institute for Microbial Diseases, Osaka University, Suita, Japan.
(16)Division of Molecular Design, Medical Institute of Bioregulation, Kyushu University, Fukuoka, Japan.
(17)Division of Molecular Immunology, Medical Mycology Research Center, Chiba University, Chiba, Japan.
[Publication]
Nature Communications
Contact for this matter
Hyogo College Hyogo Medical University General Affairs Department Public Relations Division
TEL: 0798-45-6655 (direct)
Email: kouhou@hyo-med.ac.jp